Spectral clustering is an approach to clustering data in which the spectrum (eigenvalues) of a similarity matrix is used to perform dimension reduction. Given a symmetric similarity or adjacency matrix, \(A\). The Laplacian matrix is equal to the
\(L=D-A\) where \(D\) is equal to the degree matrix.
- Input: Similarity matrix, \(S \in \mathbb{R}^{n \times n}\), number k of clusters to construct.
- Construct a similarity graph by one of the ways described in Section 2. Let W be its weighted adjacency matrix.
- Compute the unnormalized Laplacian L.
- Compute the first k eigenvectors $u_1,\dots,u_k$ of L
- Let \(U \in \mathbb{R}^{n\times k}\) be the matrix containing the vectors \(u_1,\dots,u_k\) as columns.
- For \(i= 1,\dots,n\), let \(y_i\in \mathbb{R}^k\) be the vector corresponding to the i-th row of U. Cluster the points \((yi) i=1,\dots n\) in \(\mathbb{R}^k\) with the k-means algorithm into clusters \(C_1,\dots ,C_k\)
- Output: Clusters \(A_1,\dots,A_k\) with \(A_i=\{j|y_j\in C_i\}\).
Compactness vs Connectivity
It is important to think conceptually about what you expect your data to look like when applying clustering algorithms. With spectral clustering data is projected into a lower dimensional space where it is easily seperable.
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.cluster import SpectralClustering
import nltk
import re
import numpy as np
documents=[]
#file_sample is a list of filenames ... in this case html files
for file in file_sample:
raw = open(file,'r').read()
documents.append(nltk.clean_html(raw))
#
vect=TfidfVectorizer(min_df=1,stop_words='english')
#
tfidf = vect.fit_transform(documents)
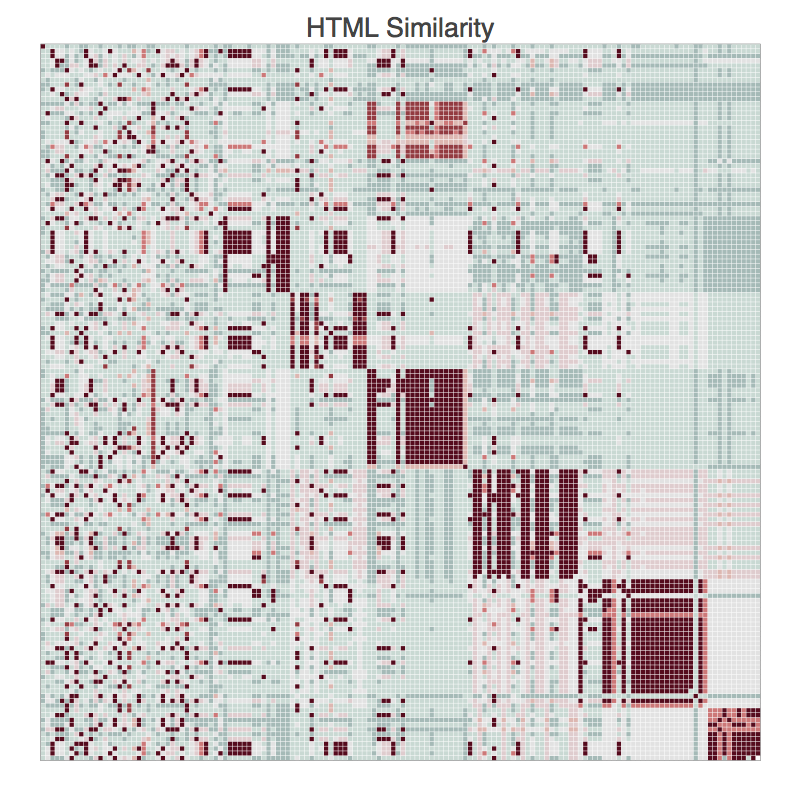
Similarity_Matrix=np.asarray((tfidf * tfidf.T).A,dtype=float)
clusters=SpectralClustering(Similarity_Matrix,affinity='precomputed',k=10)
fitted_clusters=clusters.fit_predict(Similarity_Matrix)
c=np.argsort(fitted_clusters)
sim_sort=Similarity_Matrix
for i in range(0,len(fitted_clusters)):
for j in range(0,len(fitted_clusters)):
sim_sort[i,j]=Similarity_Matrix[c[i],c[j]]